Publications
Highlights
(For a full list see below or go to Google Scholar, dblp (CS only))
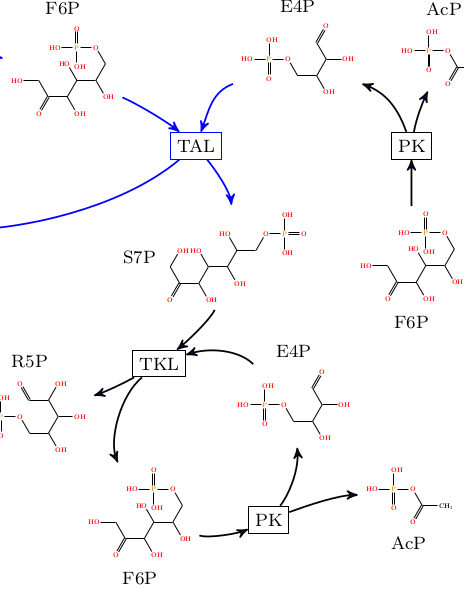
We present an elaborate framework for formally modelling pathways in chemical reaction networks on a mechanistic level. Networks are modelled mathematically as directed multi-hypergraphs, with vertices corresponding to molecules and hyperedges to reactions. Pathways are modelled as integer hyperflows and we expand the network model by detailed routing constraints.
Jakob L. Andersen, Christoph Flamm, Daniel Merkle, Peter F. Stadler
IEEE/ACM Transactions on Computational Biology and Bioinformatics 16:2, 2019
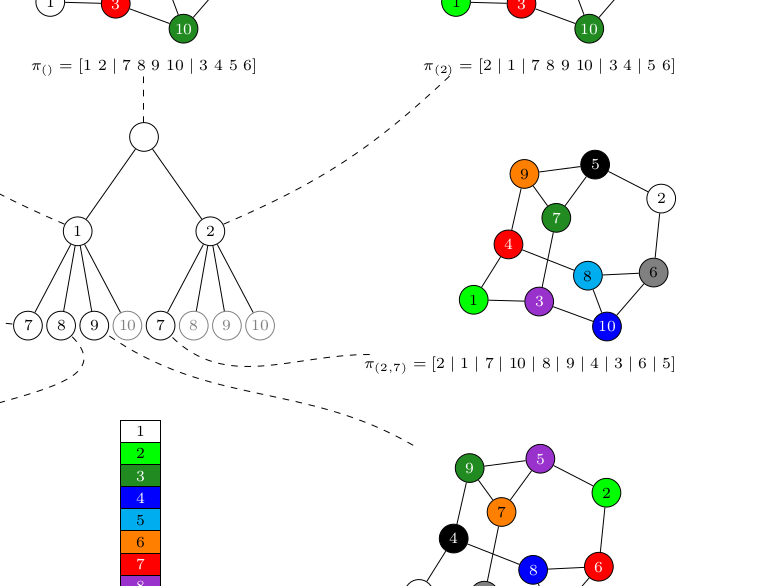
We present an algorithmic software framework that facilitates implementation of heuristics as independent extensions to a common core algorithm. Using collections of different graph classes we investigate the effect of varying the selections of heuristics, often revealing exactly which individual algorithmic choice is responsible for particularly good or bad performance. On several benchmark collections, including a newly proposed class of difficult instances, we additionally find that our implementation performs better than the current state-of-the-art tools.
Jakob L. Andersen, Daniel Merkle
2018 Proceedings of the Twentieth Workshop on Algorithm Engineering and Experiments (ALENEX)
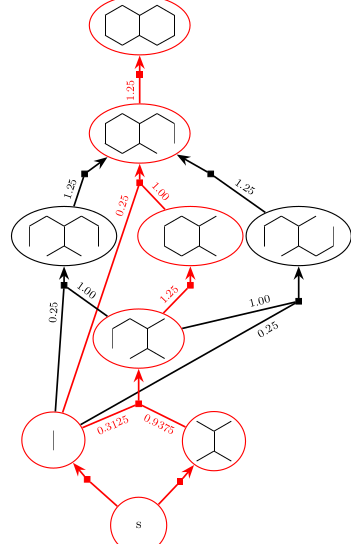
We demonstrate that synthesis planning can be phrased as a combinatorial optimization problem on hypergraphs by modeling individual synthesis plans as directed hyperpaths embedded in a hypergraph of reactions (HoR) representing the chemistry of interest. As a consequence, a polynomial time algorithm to find the K shortest hyperpaths can be used to compute the K best synthesis plans for a given target molecule.
Rolf Fagerberg, Christoph Flamm, Rojin Kianian, Daniel Merkle and Peter F. Stadler
Journal of Cheminformatics 2018, 10:19
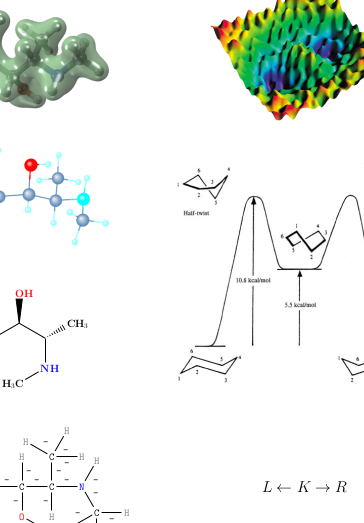
The formalization of chemical reactions as graph grammars provides a generative system, well grounded in category theory, at the right level of abstraction for the analysis of large and complex reaction networks. We argue that the intermediate-level theory outlined here holds promise in many fields of chemistry. In particular, we suggest that it is a plausible substrate for a predictive theory of prebiotic chemistry.
Jakob L. Andersen, Christoph Flamm, Daniel Merkle, Peter F. Stadler
References for highlights
Chemical Transformation Motifs—Modelling Pathways as Integer Hyperflows
Jakob L. Andersen, Christoph Flamm, Daniel Merkle, Peter F. Stadler
IEEE/ACM Transactions on Computational Biology and Bioinformatics 16:2, 2019
A Generic Framework for Engineering Graph Canonization Algorithms
Jakob L. Andersen, Daniel Merkle
2018 Proceedings of the Twentieth Workshop on Algorithm Engineering and Experiments (ALENEX)
Finding the K best synthesis plans
Rolf Fagerberg, Christoph Flamm, Rojin Kianian, Daniel Merkle and Peter F. Stadler
Journal of Cheminformatics 2018, 10:19
An Intermediate Level of Abstraction for Computational Systems Chemistry
Jakob L. Andersen, Christoph Flamm, Daniel Merkle, Peter F. Stadler
Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 2017
Peer-reviewed journal articles (Jan 2021)
-
Nøjgaard N., W. Fontana, M. Hellmuth, and Daniel Merkle: Cayley Graphs of Semigroups Applied to Atom Tracking in Chemistry. Journal of Computational Biology. accepted
-
Flamm, C., M. Hellmuth, D. Merkle, N. Nøjgaard, & P.F. Stadler (2020). Generic Context-Aware Group Contributions. IEEE/ACM Transactions on Computational Biology and Bioinformatics. (early access: https://doi.org/10.1109/tcbb.2020.2998948)
-
Andersen, J.L., C. Flamm, D. Merkle, P.F. Stadler. Defining Autocatalysis in Chemical Reaction Networks. Journal of Systems Chemistry, 8, 2020
-
Andersen, J. L. and D. Merkle: A Generic Framework for Engineering Graph Canonicalization Algorithms. ACM Journal of Experimental Algorithmics (JEA), 25(1), 2020.
-
Andersen, J. L., D. Merkle and P. Severin: Combining Graph Transformations and Semigroups for Isotopic Labeling Design. Journal of Computational Biology, 27(2):269–287, 2020.
-
Andersen, J. L., C. Flamm, D. Merkle, P. F. Stadler: Chemical Transformation Motifs — Modelling Pathways as Integer Hyperflows. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 16:2, 510–523, 2019.
-
Hansen, Bjarke N., Kim S. Larsen, Daniel Merkle, Alexei Mihalchuk: DNA-Templated Synthesis Optimization. Natural Computing, 17(4): 693-707, 2018.
-
Fagerberg, R., C. Flamm, R. Kianian, D. Merkle, P.F. Stadler: Finding the K Best Synthesis Plans. Journal of Cheminformatics, 10:19, 2018.
-
Nøjgaard, Nikolai, Manuela Geiß, Daniel Merkle, Peter F. Stadler, Nicolas Wieseke, Marc Hellmuth: Time-consistent reconciliation maps and forbidden time travel. Algorithms for Molecular Biology, 13(1):2:1–2:17, 2018.
-
Andersen, J .L., R. Fagerberg, C. Flamm, R. Kianian, D. Merkle, P. F. Stadler: Towards Mechanistic Prediction of Mass Spectra Using Graph Transformation. MATCH Communications in Mathematical and in Computer Chemistry, 80(3):705–731, 2018.
-
Andersen, J. L., C.Flamm, D. Merkle, P. F. Stadler: Rule Composition in Graph Transformation Models of Chemical Reactions. MATCH Communications in Mathematical and in Computer Chemistry, 80(3):661–704, 2018.
-
Andersen, J. L., C. Flamm, D. Merkle, Peter F. Stadler: An Intermediate Level of Abstraction for Computational Systems Chemistry. Philosophical Transactions of the Royal Society of London A: Mathematical, Physical and Engineering Sciences, 375(2109), 2017.
-
Andersen, J. L., C. Flamm, D. Merkle, P. F. Stadler: In silico Support for Eschenmoser’s Glyoxylate Scenario. Israel Journal of Chemistry, 55(8):919–933, 2015.
-
Merkle, D., M. Middendorf, A. Scheidler: Task Allocation in Organic Computing Systems: Networks with Reconfigurable Helper Units. International Journal of Autonomous and Adaptive Communications Systems, 8(1):60–80, 2015.
-
Andersen, J. L., C. Flamm, D. Merkle, P. F. Stadler: Generic Strategies for Chemical Space Exploration. International Journal of Computational Biology and Drug Design, 7((2/3)):225–258, 2014.
-
Andersen, J. L., T. Andersen, C. Flamm, M. M. Hanczyc, D. Merkle, P. F. Stadler: Navigating the Chemical Space of HCN Polymerization and Hydrolysis: Guiding Graph Grammars by Mass Spectrometry Data. Entropy, 15(10):4066–4083, 2013.
-
Fagerberg, R., C. Flamm, D. Merkle, P. Peters, P.F. Stadler: On the Complexity of Reconstructing Chemical Reaction Networks. Mathematics in Computer Science, 7(3): 275–292, 2013.
-
Andersen, J. L., C. Flamm, D. Merkle, Peter F. Stadler: Inferring Chemical Reaction Patterns Using Rule Composition in Graph Grammars. Journal of System Chemistry, 4(4), 2013.
-
Scheidler, A. D. Merkle, M. Middendorf: Swarm Controlled Emergence for Ant Clustering. International Journal of Intelligent Computing and Cybernetics, 6(1):62–82, 2013.
-
Andersen, J.L., C. Flamm, D. Merkle, P.F. Stadler: Maximizing Output and Recognizing Autocatalysis in Chemical Reaction Networks is NP-Complete. Journal of System Chemistry, 3(1), 2012.
-
Bernt, M., K.-Y. K.Y. Chen, M.C. Chen, A.-C. Chu, D. Merkle, H.-L. Wang, K.-M. Chao,
M. Middendorf: Finding All Sorting Tandem Duplication Random Loss Operations. Journal of Discrete Algorithms, 9(1), 2011. -
Merkle, D., Martin Middendorf, Nicolas Wieseke: A Parameter-Adaptive Dynamic Programming Approach for Inferring Cophylogenies. BMC Bioinformatics, 11(S-1):60, 2010.
-
Hellmuth, M., D. Merkle, M. Middendorf: Extended Shapes for the Combinatorial Design of RNA Sequences. International Journal of Computational Biology and Drug Design, 2(4):371–384, 2009.
-
Diwold, K., D. Merkle, M. Middendorf: Adapting to Dynamic Environments: Polyethism in Response Threshold Models for Social Insects. Advances in Complex Systems, 12(3):327–346, 2009.
-
Bernt, M., D. Merkle, M. Middendorf: Solving the Preserving Reversal Median Problem. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 5(3): 332-347, 2008.
-
Perseke, M., Fritzsch, G., K. Ramsch, M. Bernt, D. Merkle, M. Middendorf, D. Bernhard, P.F. Stadler, M. Schlegel: Phylogenetic Analyses of Mitochondrial Genomes of Echinoderms Challenge the Monophyly of Eleutherozoa. Molecular Phylogenetics and Evolution, 47: 855–864, 2008.
-
Merkle, D., M. Middendorf: Swarm Intelligence and Signal Processing. IEEE Signal Processing Magazine, 25(6): 152-158, 2008.
-
Thalheim, T., D. Merkle, M. Middendorf: Protein Folding in the HP-Model Solved With a Hybrid Population Based ACO Algorithm. IAENG International Journal of Computer Science, 35(3): 291-300, 2008.
-
Janson, S., D. Merkle, M. Middendorf:: A Decentralization Approach for Swarm Intelligence Algorithms in Networks Applied to Multi Swarm PSO. International Journal of Intelligent Computing and Cybernetics, 1(1):24–45, 2008. .
-
Janson, S., D. Merkle, M. Middendorf: Molecular Docking with Multi-Objective Particle Swarm Optimization. Applied Soft Computing, 8(1):666–675, 2008.
-
Scheidler, A., D. Merkle, M. Middendorf: Stability and Performance of Ant Queue Inspired Task Division Methods. Theory in Biosciences, 127(2):149–161, 2008.
-
Merkle, D., M. Middendorf, A. Scheidler: Self-Organized Task Allocation for Service Tasks in Computing Systems with Reconfigurable Components. Journal Mathematical Modelling and Algorithms, 7(2): 237–254, 2008.
-
Bernt, M., D. Merkle, K. Ramsch, Fritzsch, G., M. Perseke, D. Bernhard, M. Schlegel, P.F. Stadler, M. Middendorf: CREx: Inferring Genomic Rearrangements Based on Common Intervals. Bioinformatics, 23(21):2957–2958, 2007.
-
Bernt, M., D. Merkle, M. Middendorf: Using Median Sets for Inferring Phylogenetic Trees. Bioinformatics, 23:e129–e135, 2007. This paper won the 3rd best paper award at the European Conference on Computational Biology 2006.
-
Bernt, M., D. Merkle, M. Middendorf: Genome Rearrangement Based on Reversals that Preserve Conserved Intervals. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 3(3): 275–288, 2007.
-
Merkle, D., M. Middendorf, A. Scheidler: Modelling Ant Brood Tending Behaviour with Cellular Automata. Journal of Cellular Automata, 2(2):183–194, 2006.
-
Merkle, D., C. Blum: Swarm Intelligence – An Optimization-based Introduction. Künstliche Intelligenz, 4:5–10, 2005.
-
Merkle, D., M. Middendorf: Reconstruction of the Cophylogenetic History of Related Phylogenetic Tree with Divergence Timing Information. Theory in Biosciences, 123(4):277–299, 2005.
-
Merkle, D., M. Middendorf, A. Scheidler: Decentralized Packet Clustering in Router-based Networks. International Journal of Foundations of Computer Science, 16(2):321–341, 2005.
-
Merkle, D., M. Middendorf: On Solving Permutation Scheduling Problems with Ant Colony Optimization. International Journal of Systems Science, 36(5):255–266, 2005.
-
Merkle, D., M. Middendorf: Dynamic Polyethism and Competition for Tasks in Threshold Reinforcement Models of Social Insects. Adaptive Behavior, 12:251–262, 2004.
-
Janson, S., D. Merkle, M. Middendorf, H. ElGindy, H. Schmeck: On Enforced Convergence of ACO and its Implementation on the Reconfigurable Mesh Architecture Using Size Reduction Tasks. Journal of Supercomuting, 26(3): 221-238,2003.
-
Merkle, D., M. Middendorf: Modelling the Dynamics of Ant Colony Optimization. Evolutionary Computation, 10(3):235–265, 2002.
-
Merkle, D., M. Middendorf, H. Schmeck: Ant Colony Optimization for Resource-Constrained Project Scheduling. IEEE Transactions on Evolutionary Computation 6(4):333–346, 2002.
-
Merkle, D., M. Middendorf: Fast Ant Colony Optimization on Runtime Reconfigurable Processor Arrays. Genetic Programming and Evolvable Machines, 3(4):345–361, 2002.
-
Merkle, D., Th. Worsch , Formal language recognition by stochastic cellular automata. Fundamenta Informaticae, 52(1-3):183–201, 2002.
- Merkle, D., M. Middendorf: An Ant Algorithm with Global
Pheromone Evaluation for Scheduling a Single Machine. Applied
Intelligence, 18(1):105–111, 2002.
Peer-reviewed conference articles
-
Hellmuth, M., D. Merkle and N. Nøjgaard: Atom Tracking Using Cayley Graphs. In 16th International Symposium on Bioinformatics Research and Applications (ISBRA), Lecture Notes in Computer Science, vol 12304, 406-415, 2020.
-
Andersen J.L., M. Hellmuth, D. Merkle, N. Nøjgaard, M. Peressotti. A Graph-Based Tool to Embed the $\pi$-Calculus into a Computational DPO Framework. In Proceedings of the SOFSEM 2020 Doctoral Student Research Forum, CEUR Workshop Proceedings, 121-132, 2020
-
Andersen, J. L., Daniel Merkle, Peter Severin: Graph Transformations, Semigroups, and Isotopic Labeling. 15th International Symposium on Bioinformatics Research and Applications (ISBRA), Proceedings, Lecture Notes in Computer Science, vol 11490, 196-207, 2019.
-
Nøjgaard, Nikolai, Nadia El-Mabrouk, Daniel Merkle, Nicolas Wieseke, Marc Hellmuth: Partial Homology Relations - Satisfiability in Terms of Di-Cographs. Computing and Combinatorics - 24th International Conference, COCOON , Proceedings, 10976 Lecture Notes in Computer Science, 403–415. Springer, 2018.
-
Andersen, J. L., D. Merkle: A Generic Framework for Engineering Graph Canonicalization Algorithms. Proceedings of the Twentieth Workshop on Algorithm Engineering and Experiments, ALENEX 2018. SIAM, 139–153. 2018.
-
Hellmuth, M., A.S. Knudsen, M. Kotrbčík, D. Merkle, N. Nøjgaard: Linear Time Canonicalization and Enumeration of Non-Isomorphic 1-Face Embeddings. Proceedings of the Twentieth Workshop on Algorithm Engineering and Experiments, ALENEX 2018. SIAM, 2018. 154-168. 2018.
-
Hansen, B. N., K. S. Larsen, D. Merkle, Alexei Mihalchuk: DNA-Templated Synthesis Optimization. Proceedings of the 23rd International Conference on DNA Computing and Molecular Programming - DNA 23, 10467 Lecture Notes in Computer Science, 17–32. Springer, 2017.
-
Nøjgaard, N., M. Geiß, D. Merkle, P. F. Stadler, N. Wieseke, M. Hellmuth: Forbidden Time Travel: Characterization of Time-Consistent Tree Reconciliation Maps. 17th International Workshop on Algorithms in Bioinformatics, WABI, 88 LIPIcs, 17:1–17:12. Schloss Dagstuhl - Leibniz-Zentrum fuer Informatik, 2017.
-
Andersen, J. L., C. Flamm, D. Merkle, P. F. Stadler: Chemical Graph Transformation with Stereo-Information. Proceedings of the 10th International Conference on Graph Transformation, ICGT 2017, 10373 Lecture Notes in Computer Science, 54–69. Springer, 2017.
-
Andersen, J. L., C. Flamm, D. Merkle, P. F. Stadler: A Software Package for Chemically Inspired Graph Transformation. Graph Transformation - 9th International Conference (ICGT 2016), 9761 Lecture Notes in Computer Science, 73–88. Springer, 2016.
-
Flamm, C., D. Merkle, P. F. Stadler, U. Thorsen: Automatic Inference of Graph Transformation Rules Using the Cyclic Nature of Chemical Reactions. Graph Transformation - 9th International Conference (ICGT 2016), 9761 Lecture Notes in Computer Science, 206–222. Springer, 2016.
-
Andersen, J. L., C. Flamm, D. Merkle, P. F. Stadler: 50 Shades of Rule Composition - From Chemical Reactions to Higher Levels of Abstraction. Proceedings of the 1st International Conference on Formal Methods in Macro-Biology (FMMB), 8738 Lecture Notes in Computer Science, 117–135. Springer, 2014.
-
Hargreaves, F., D. Merkle, P. Schneider-Kamp: Group Communication Patterns for High Performance Computing in Scala. Proceedings of the 3rd ACM SIGPLAN Workshop on Functional High-Performance Computing (FHPC), 75–85, 2014.
-
Andersen, J. L., C. Flamm, Hanczyc M.M., D. Merkle: Towards an Optimal DNA-Templated Molecular Assembler. Proceedings of 14th International Conference on Artificial Life (ALIFE)*, 557–564. MIT Press, 2014.
-
Hargreaves, F. P., D. Merkle: FooPar: A Functional Object Oriented Parallel Framework in Scala. Proc. of the 10th International Conference on Parallel Processing and Applied Mathematics (PPAM), 8385 Lecture Notes in Computer Science, 118–129. Springer, 2013.
-
Flamm, C., C. Hemmingsen, D. Merkle: Barrier Trees for Metabolic Adjustment Landscapes. Proc. of the 12th European Conference on Artificial Life (ALIFE), 1151–1158, 2013.
-
Fagerberg, R., C. Flamm, D. Merkle,
P. Peters: Exploring Chemistry Using SMT. Proceeding of the 18th International Conference on Principles and Practice of Constraint Programming (CP 2012), 7514 Lecture Notes in Computer Science, 900–915. Springer, 2012. -
Bernt, M, M.-C. Chen, D. Merkle, H.-L. Wang, K.-M. Chao, M. Middendorf: Proc. 2009 of the 20th Annual Symposium on Combinatorial Pattern Matching (CPM 2009), 5577 Lecture Notes in Computer Science (LNCS), 301–313, Springer, Berlin, 2009
-
Hellmuth, M., D. Merkle, M. Middendorf: Proceedings of the International Joint Conferences on System Biology, Bioinformatics and Intelligent Computing (IJCBS 2009), Shanghai, China, 2009.
-
Diwold, K., D. Merkle, M. Middendorf: The Influence of Dynamic Environments on Polyethism in Response Threshold Models for Social Insects. Proc. of the 8th German Workshop on Artificial Life (GWAL), 37–48. IOS Press, 2009.
-
Bernt, M., D. Merkle, M. Middendorf: An Algorithm for Inferring Mitochondrial Genome Rearrangements in a Phylogenetic Tree. Proc. 2008 of the 6th RECOMB Comparative Genomics Satellite Workshop (RECOMB-CG), 5267 Lecture Notes in Bioinformatics (LNBI), 143–157, Springer, Berlin, 2008.
-
Thalheim, T., D. Merkle, M. Middendorf: A Hybrid Population based ACO Algorithm for Protein Folding. Proc. 2008 IAENG International Conference on Bioinformatics (ICB), 2008.
-
Brutschy, A., A. Scheidler, D. Merkle, M. Middendorf: Learning from House-Hunting Ants: Collective Decision-Making in Organic Computing Systems. Proc. of the 6th International Conference on Ant Colony Optimization and Swarm Intelligence (ANTS 2008), 5217 Lecture Notes in Computer Science (LNCS), 96–107. Springer, Berlin, 2008. This paper won the paper award at ANTS 2008.
-
Scheidler, A., C. Blum, D. Merkle, M. Middendorf: Emergent Sorting in Networks of Router Agents. Proc. of the 6th International Conference on Ant Colony Optimization and Swarm Intelligence (ANTS 2008), 5217 Lecture Notes in Computer Science (LNCS), 299–306. Springer, Berlin, 2008.
-
Scheidler, A., D. Merkle, M. Middendorf: Congestion Control in Ant Like Moving Agent Systems. 2nd IFIP Conference on Biologically Inspired Collaborative Computing (BICC), 2008.
-
Merkle, D., M. Middendorf, A. Scheidler: Swarm Controlled Emergence - Designing an Anti-Clustering Ant System. Proc. of the IEEE Swarm Intelligence Symposium 2007 (SIS), 242–249. IEEE Press, 2007.
-
Bernt, M., D. Merkle, M. Middendorf: A Fast and Exact Algorithm for the Perfect Reversal Median Problem.
Bioinformatics Research and Applications (ISBRA), 4463 Lecture Notes in Bioinformatics (LNBI), 305–316, Springer, Berlin, 2007. -
Scheidler, A., D. Merkle, M. Middendorf: Stability and Performance of Ant Queue Inspired Task Division Methods. Proc. of the European Conference on Complex Systems (ECCS), 2007.
-
Bernt, M., D. Merkle, M. Middendorf: The Reversal Median Problem, Common Intervals, and Mitochondrial Gene Orders. Computational Life Sciences II - Proc. 2nd International Symposium (CompLife), 4216 Lecture Notes in Bioinformatics (LNBI), 52–63, 2006.
-
Merkle, D., M. Middendorf, A. Scheidler: Using Decentralized Clustering for Task Allocation in Networks with Reconfigurable Helper Units. Proceedings of the International Workshop on Self-Organizing Systems 2006 (IWSOS 2006), 4124 Lecture Notes in Computer Science (LNCS), 137–147. Springer, Berlin, 2006.
-
Merkle, D., M. Middendorf, A. Scheidler: Self-Organized Task Allocation for Computing Systems with Reconfigurable Components. Proceedings of the 9th International Workshop on Nature Inspired Distributed Computing (NIDISC’06), 2006.
-
Scheidler, A., D. Merkle, M. Middendorf: Emergent Sorting Patterns and Individual Differences of Randomly Moving Ant Like Agents. Proceedings of the 7th German Workshop on Artificial Life (GWAL-7), 2006.
-
Bernt, M., D. Merkle, M. Middendorf: A Parallel Algorithm for Solving the Reversal Median Problem. Proceedings of the Parallel Processing and Applied Mathematics - Bio-Computing Workshop (PBC 2005), 3911 Lecture Notes in Computer Science (LNCS), 1089–1096, Springer, Berlin, 2006.
-
Merkle, D., M. Middendorf, A. Scheidler: Dynamic Decentralized Packet Clustering in Networks.
Proceedings of the 2nd European Workshop on Evolutionary Algorithms in Stochastic and Dynamic Environments (EVOSTOC), 3449 Lecture Notes in Computer Science (LNCS), 574–583, Springer, Berlin, 2005. -
Janson, S., D. Merkle, M. Middendorf: Decentralized Packet Clustering in Networks Applied to Particle Swarm Optimization. Proceedings of the Conference on Design, Analysis, and Simulation of Distributed Systems 2005 (DASD 2005), 3–10, 2005.
-
Janson, S., D. Merkle: A New Multi-objective Particle Swarm Optimization Algorithm Using Clustering Applied to Automated Docking. Proc. of the Second International Workshop on Hybrid Metaheuristics (HM), 3636 Lecture Notes in Computer Science (LNCS), 128–141, 2005.
-
Merkle, D., M. Middendorf, A. Scheidler: Modelling Ant Brood Tending with Cellular Automata. Proceedings of the Workshop on Modelling of Complex Systems by Cellular Automata 2005 (ICCS 2005), 3515 Lecture Notes in Computer Science (LNCS), 412–419. Springer, Berlin, 2005.
-
Merkle, D., M. Middendorf: Ant Colony Optimization: Biological Motivation, Phase Structure, and Modelling. Proceedings of the first International Symposium on Mathematical and Computational Biology (BIOMAT), 2005.
-
Merkle, D., M. Middendorf, A. Scheidler: Decentralized Packet Clustering in Networks. Proceedings of the International Parallel and Distributed Processing Symposium (IPDPS-2004), Sixth International Workshop on Nature Inspired Distributed Computing (NIDISC’04). IEEE-CS Press, 9 pages, 2004.
-
Merkle, D., M. Middendorf: Competition Controlled Pheromone Update for Ant Colony Optimization. Proceedings of the Fourth International Workshop on Ant Colony and Swarm Intelligence (ANTS 2004), 3172 Lecture Notes in Computer Science (LNCS), 95–105, Springer, Berlin, 2004.
-
Merkle, D., M. Middendorf: Dynamic Polyethism in Social Insect Societies - A Simulation Study. Proceedings of the 2nd International Workshop on the Mathematics and Algorithms of Social Insects, 99–106, Georgia Institute of Technology, Atlanta, 2003.
-
Merkle, D., M. Middendorf: Studies on the dynamics of ant colony optimization algorithms. Proceedings of the Genetic and Evolutionary Computation Conference (GECCO-2002), 105–112, San Francisco, CA, Morgan Kaufmann, USA, 2002.
-
Merkle, D., M. Middendorf: Modelling ACO: Composed permutation problems. Proceedings of 3rd International Workshop on Ant Algorithms (ANTS’2002), 2463 Lecture Notes in Computer Science (LNCS), 149–162, Springer, Berlin, 2002.
-
Merkle, D., M. Middendorf: Ant colony optimization with the relative pheromone evaluation method. Proceedings of the 3rd European Workshop on Scheduling and Timetabling and 3rd European Workshop on Evolutionary Methods for AI Planning (EvoSTIM/EvoPLAN), 2279 Lecture Notes in Computer Science (LNCS), 325–333, 2002.
-
Branke, J., M. Decker, D. Merkle: Coevolutionary Ant Algorithms for Playing Games. Proceedings of 3rd International Workshop on Ant Algorithms (ANTS’2002), 2463 Lecture Notes in Computer Science (LNCS), 298–299, Springer, Berlin, 2002.
-
Janson, S., D. Merkle, M. Middendorf, H. ElGindy, H. Schmeck: On Enforced Convergence of ACO and its Implementation on the Reconfigurable Mesh Architecture Using Size Reduction Tasks. Proceedings of the Second International Conference on Engineering of Reconfigurable Systems and Algorithms (ERSA’02), 3–9, 2002.
-
Iredi, S., D. Merkle, M. Middendorf: Bi-criterion Optimization with Multi Colony Ant Algorithms. , 1993 Lecture Notes in Computer Science (LNCS), 359–372, Springer, Berlin, 2001.
-
Merkle, D., M. Middendorf: A New Approach to Solve Permutation Scheduling Problems with Ant Colony Optimization. Second European Workshop on Scheduling and Timetabling (EvoStim2001), 2037 Lecture Notes in Computer Science (LNCS), 484–493. Springer, Berlin, 2001.
-
Merkle, D., M. Middendorf: Collective Optimization: The Artificial Ants Way. Abstracts of the Workshop From Worker to Colony: Understanding the Organisation of Insect Societies, Cambridge, UK, 2001. Isaac Newton Institute for Mathematical Sciences.
-
Merkle, D., M. Middendorf: Fast Ant Colony Optimization on Reconfigurable Processor Arrays. Proceedings of the 15th International Parallel and Distributed Processing Symposium (IPDPS-01). IEEE Computer Society Press, 2001.
-
Merkle, D., M. Middendorf: On the Behaviour of Ant Algorithms: Studies on Simple Problems. Proceedings of the 4th Metaheuristics International Conference (MIC’ 2001), Porto, Portugal, 573–577, 2001.
-
Merkle, D., M. Middendorf: Prospects for Dynamic Algorithm Control: Lessons from the Phase Structure of Ant Scheduling Algorithms. Proceedings of the 2001 Genetic and Evolutionary Computation Conference (GECCO) – Workshop Program, 121–126, San Francisco, CA, Morgan Kaufmann, USA, 2001.
-
Merkle, D., M. Middendorf, H. Schmeck: Ant Colony Optimization for Resource-Constrained Project Scheduling. Proceedings of the Genetic and Evolutionary Computation Conference (GECCO 2000), 893–900, San Francisco, CA, Morgan Kaufmann, USA, 2000.
-
Merkle, D., M. Middendorf: An Ant Algorithm with a New Pheromone Evaluation Rule for Total Tardiness Problems. Proceeding of the EvoWorkshops 2000, 1803 Lecture Notes in Computer Science (LNCS), 287–296. Springer, Berlin, 2000.
-
Merkle, D., M. Middendorf, H. Schmeck: Ant Colony Optimization for the RCPSP using Weighted Summation Evaluation. From Ant Colonies to Artificial Ants: Second International Workshop on Ant Algorithms (ANTS), Proceedings, 84–87, 2000.
- Merkle, D., M. Middendorf, H. Schmeck:
Pheromone Evaluation in Ant Colony Optimization.
Proceedings of the 26th Annual Conference on Industrial
Electronics, Control and Instrumentation
(IECON-2000), 2726–2731, IEEE Press, Piscataway, USA, 2000
Editorials
-
Andersen, Jakob L., C. Flamm, D. Merkle, P.F. Stadler: Report from Dagstuhl Seminar 17452: Algorithmic Cheminformatics, 2018.
-
Banzhaf, W., C. Flamm, D. Merkle, P.F. Stadler : Report from Dagstuhl Seminar 14452: Algorithmic Cheminformatics, 2015.
-
Klemm, K., D. Merkle, E. Olbrich: Editorial: Artificial Life. Advances in Complex Systems, 12(3), 2009.
- Doerner, K.F., D. Merkle, T. Stützle:
Special Issue on Ant Colony Optimization. Swarm Intelligence,
3(1):1–2, 2009.
Books (edited)
-
Blum, C., D. Merkle: 群智能 : 介绍与应用 (Swarm Intelligence - Introduction and Application, Chinese Translation). Chinese publisher: National Defense Industry Press with permission from Springer, 2011.
-
Fellermann, H., Mark Dörr, M. Hanczyc, L. Laursen, S. Maurer, D. Merkle, P.-A. Monnard, K. Stoy, S. Rasmussen : Proceedings of the 12th International Conference on the Synthesis and Simulation of Living Systems, ALife XII (University of Southern Denmark). MIT Press, 2010.
-
Klemm, K., D. Merkle, E. Olbrich : Proceedings of the 8th German Workshop on Artificial Life. IOS Press, 2009.
- Blum, C., D. Merkle : Swarm Intelligence -
Introduction and Application. Natural Computing Series, Springer,
2008.
Book chapters
-
Andersen, Jakob Lykke, Christoph Flamm, Daniel Merkle, Peter F. Stadler: Information Processing in Chemical Systems. al., David H. Wolpert et : The Interplay of Thermodynamics and Computation in Natural and Artificial Systems. The Santa Fe Institute Press, 2019. accepted.
-
Bernt, M., D. Merkle, M. Middendorf, B. Schierwater, M. Schlegel, P. F. Stadler: Computational Methods for the Analysis of Mitochondrial Genome Rearrangements, Deep Metatzoan Phylogeny The Backbone of the Tree of Life, 515–529. De Gruyter, 2014.
-
Merkle, D., M. Middendorf: Swarm Intelligence. Burke, E. G. Kendall : Search Methodologies – Introductory Tutorials in Optimisation and Decision Support Methodology. Springer, 2014.
-
Scheidler, A., A. Brutschy, K. Diwold, D. Merkle, M. Middendorf: Organic Computing: A Paradigm Shift for Complex Systems, Ant Inspired Methods for Organic Computing, 95–110. Springer, 2011.
-
Merkle, D., M. Middendorf, A. Scheidler: Organic Computing and Swarm Intelligence. Blum, C.
D. Merkle : Swarm Intelligence, 253–281. Springer, 2008. -
Merkle, D., M. Middendorf, A. Scheidler: Organic Computing, Self-Adaptive Worker-Helper Systems with Self-Organized Task Allocation, Würtz, R.P. (editor): Organic Computing, 221–240. Springer, 2008.
-
Petzold, E., A. von Haeseler, D. Merkle, M. Middendorf, H.A. Schmidt: Phylogenetic Parameter Estimation on COWs. Zomaya, A. : Parallel Computing in Bioinformatics and Computational Biology, 347–368. Wiley, 2006.
-
Merkle, D., M. Middendorf: Swarm Intelligence. Burke, E. G. Kendall : Introductory Tutorials in Optimisation, Search and Decision Support Methodology, 401–435. Springer, 2005.
-
Janson, S., D. Merkle, M. Middendorf: Parallel Ant Colony Algorithms. Alba, E. : Parallel Metaheuristics, Wiley Book Series on Parallel and Distributed Computing, 171 – 201. Wiley, 2005.
- Merkle, D., M. Middendorf: On the Behaviour of Ant
Algorithms: Studies on Simple Problems. Resende, M.G.C.
J.P. de Sousa : Metaheuristics: Computer Decision-Making. pages 465-480, Kluwer Academic Publisher, 2003.
Book review
- Merkle, D., M. Middendorf: Book Review on “Ant Colony Optimization” by M. Dorigo, T. Stützle. European Journal of Operations Research, 168(1):269–271, 2005.
