DM813 Mandatory Assignment 2: An ILP formulation for fixing estimated orthology matrices
Biological Background
The reconstruction of the evolutionary history of a gene family is necessarily based on at least three interrelated types of information. The true phylogeny of the investigated species is required as a scaffold with which the associated gene tree must be reconcilable. On the other hand, orthology or paralogy of genes found in different species determines whether an internal vertex in the gene tree corresponds to a duplication or a speciation event. The latter, in turn, are reflected in the species tree. The reconciliation of gene and species trees is a very widely studied problem. In most practical applications, however, neither the gene tree nor the species tree can be determined unambiguously. Although orthology information is often derived from the reconciliation of a gene tree with a species tree, recent benchmarks have shown that orthology can also be inferred at similar levels of accuracy without the need to construct trees by means of clustering-based approaches such as ProteinOrtho [4]. In [1] the following question was addressed: How much information about the gene tree, the species tree, and their reconciliation is already contained in the orthology relation between genes? According to the definition of Fitch [2], two genes are (co-)orthologous if their last common ancestor in the gene tree represents a speciation event. Otherwise, i.e., when their last common ancestor is a duplication event, they are paralogs. The orthology relation on a set of genes is therefore determined by the gene tree T and an "event labeling" that identifies each interior vertex of T as either a duplication or a speciation event. One of the main results of [1], which relies on the theory of symbolic ultrametrics developed in [3], is the following: A relation on a set of genes is an orthology relation (i.e. it derives from some event-labeled gene tree) if and only if it is a cograph, i.e. it is not allowed to have a path of length 4 as an induced subgraph. This property will be used for solving the subproblem (1) below.
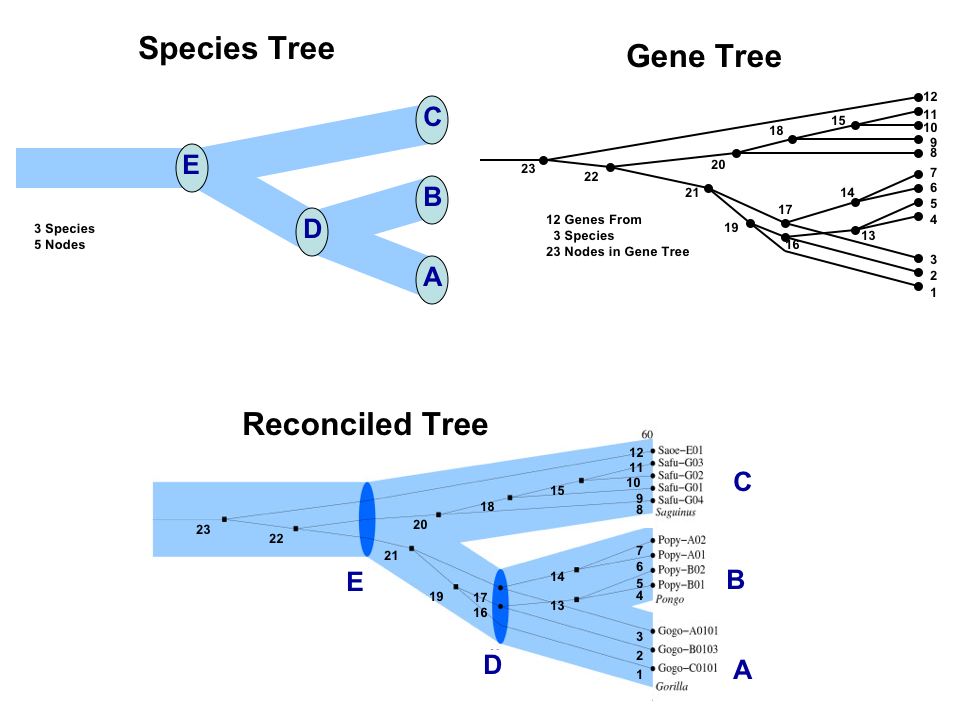
The following Figure (from here) is an example of a species tree, a gene tree, and a reconciled tree. Events 16 and 17 are speciation events, events 13, 14, 15, 18, 19, 20, 21, 22, and 23 are duplication events.
Prerequisites
In this assignment you are given an estimated orthology matrix, and
your task is to fix the inconsistencies that occur in the matrix with
an Integer
Linear Programming (ILP) approach. Please make sure you understand
the slide set week47-nic-ILP.pdf properly before you work
on this problem. The slide-set can be found in the Blackboard
system. Nicolas Wieseke provided the idea and the source code
templates for this assignment.
Your task
Your task will be to fix the estimated orthology matrix in an stepwise fashion.
- (1) In the first step the estimated orthology matrix has to be made gene-tree-reconcilable, i.e., (formulated as a graph theoretical problem) the orthology relation sought seen as a undirected graph is not allowed to have a path of length 4 as an induced subgraph, and it should be created based on a minimal number of changes in the estimated orthology matrix. Note, that this step is solved already and the source code is provided.
- (2) In the second step additional constraints need to be added such that the orthology relation sought should be species-tree-reconcilable for a given species tree.
- (3) In the third step you are not given a species tree. Instead you are supposed to find a species-tree-reconcilable orthology relation for a species tree. The species tree for which the orthology map is species-tree-reconcilable has to be given as a set of rooted triples (AB|C), i.e., phylogenetic trees on three leaves with precisely two interior vertices. (Note that this set of triples can be used as input to a polynomial time algorithm to infer the phylogenetic tree, see for example [5])
What is provided
We provide you (either in blackboard, or in the directory /home/daniel/dm813 in the IMADA terminal room):
- the ILP formulation for the (sub-)problems (1)-(3) to be solved. The document is
called
assign2-ILP.pdf. - the Java source code to solve problem (1) with the constraints from the ILP formulation. We provide four version of the Java source code as we implemented solving problem (1) using three use three different solvers, namely CPLEX v12.5, GUROBI, LPSolve, and GLPK. We recommend using CPLEX to approach this problem. The source code should work on Linux, Windows, and Mac on 32bit as well as on 64bit machines. Note that you are not required to use the Java source code but you can use any programming language of your choice.
- The test instances provided are of the following format:
The k-th row (and column) corresponds to the k-th entry in the list of genes on the species. Note, that the specification is not completely consistent to the ILP formulation provided. Methods to read files of this kind are provided, too (see source code).
#phylogenetic tree ((D,E),F); #genes on species D D E E E E E D D D F F #Estimated Orthology Matrix 0 0 1 1 1 1 0 0 0 0 1 1 0 0 0 1 0 1 1 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 0 1 1 0 1 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 0 0 0 0 0 0 1 1 1 1 1 1 1 1 1 0 1 1 0 0 1 1 1 1 1 1 0 1 1 1 0 0
Report / Submission
This assignment can be done in groups of two people. The submission of the source code has to be supplemented by a write-up (max. 12 pages). The report has to be written in English. The write-up should contain in any case at least:
- A title page (title, names, date, and so on).
- An introduction to the problem including the necessary formal definitions.
- Description of approaches followed and ideas on how to optimize the implementation.
- Results on provided test instances; the test instances are called ds01, ..., ds18. For each solver you used provide a table with at least the following information for all instances:
i.) the number of changes that were needed in order to fix the estimated orthology matrix for the cases that the provided phylogenetic tree was / was not used for the matrix fixing,
ii.) the number of forbidden induced P4 subgraphs that were necessary to solve the test instance (for the cases that the provided phylogenetic tree was / was not used)
iii.) the run-time in seconds (IMADA terminal room, for the cases that the provided phylogenetic tree was / was not used),
iv.) the number of indicator split variables that are unequal to zero (only for the case that the provided phylogenetic tree was not used).
- A discussion part. The discussion part should include i.) a sub-part on what happens if you modify the equal sign of constraint 4c) of
assign2-ILP.pdfto a less-or-equal sign. This should be illustrated by a small example where the solutions are indeed different. ii.) a discussion on run-times, which includes the sizes that you were not able to solve in reasonable time (especially if you could solve all test instances). - Optional parts that could be discussed / implemented include:
- An implementation to find all optimal solutions. - A comparison of run-times of the different ILP solvers (preference should be given to CPLEX and GUROBI).
- A formal proof on the correctness of the constraints that ensure the species-tree reconiability. - A method to infer the species tree.
- The usage of callbacks in order to optimize the addition of P4 constraints.
- A statement of who contributed with what.
- A conclusion.
- References used.
Create the following directory structure on one of IMADAs pool machines
mandatory2/ mandatory2/report/ mandatory2/source/ mandatory2/results/
In the source directory there should be the (well
commented) source code of your
implementation. The results directory should contain
a README file, in which you briefly describe the tests
you made and what files can be found in the corresponding
directory. After putting all the data and the report (use pdf file
format) to the correct location, change to the
directory mandatory2 and type aflever
DM813 your@mail.com.
I would appreciate if you hand in a printout of your report and of your source code (only if you submit before Dec. 18th, the 12 pages limit applies only to the report and not to the source code). (Department secretaries office).
The strict deadline for submission (electronically) is Sunday, January, 6th, 13:00. Note however that I will only rarely read mail between Dec. 21th and Jan 3rd., consider submitting before December 21nd.
Useful References
- IBM« ILOG« CPLEX« Optimization Studio Information Center
- [1] Orthology relations, symbolic ultrametrics, and cographs, Journal of Mathematical Biology, 2012, Nicolas Wieseke Marc Hellmuth, Maribel Hernandez-Rosales, Katharina T. Huber, Vincent Moulton, Peter F. Stadler, Nicolas Wieseke
- [2] Homology: a personal view on some of the problems, Trends in Genetics, Volume 16, Issue 5, 2000, 227-231, Walter M. Fitch
- [3] Recovering Symbolically Dated, Rooted Trees from Symbolic Ultrametrics, Advances in Mathematics Volume 138, Issue 1, 1998, Pages 105-125, Sebastian B÷cker, Andreas W.M. Dress
- [4] Proteinortho: Detection of (Co-)orthologs in large-scale analysis, BMC Bioinformatics 2011, 12:124, Marcus Lechner, Sven Findei▀, Lydia Steiner, Manja Marz, Peter F Stadler and Sonja J Prohaska
- [5] Worst-case optimal approximation algorithms for maximizing triplet consistency within phylogenetic networks, Journal of Discrete Algorithms, Volume 8, Issue 1, March 2010, , 65-75, Jaroslaw Byrka, Pawel Gawrychowski, Katharina T. Huber, Steven Kelk
Frequently Asked Questions (FAQ)
- Are we allowed to work in groups?
Yes, you can work in groups. The maximal size of the group is 2 people.