DM818 Mandatory Assignment 1: Optimize Matrix Multiplication
Problem
Your task is to optimize matrix multiplication code to run fast on a single processor core of NERSC Edison cluster (please read the access instructions), and also Running Jobs on Edison. Semantics of the matrix multiplication is:
for i = 1 to n
for j = 1 to n
for k = 1 to n
C[i,j] = C[i,j] + A[i,k] * B[k,j]
end
end
end
where A, B and C are n-by-n matrices. You need to write a dgemm.c that implements this semantics and exposes it through the following C interface:
void square_dgemm( int n, double *A, double *B, double *C ).
The matrices are stored in column-major order, i.e. entry Cij is stored at C[i+j*M]. The correctness of implementation is verified using an element-wise error bound that holds for the naive matrix multiply:
|square_dgemm(n,A,B,0) - A*B| < eps * n * |A| * |B|.
This bound also holds for any other algorithm that does the same floating point operations as the naive algorithm but in different order.
However, this may exclude using some other algorithms such as Strassen's. In the formula, eps = 2–52 = 2.2 * 10–16 is the machine epsilon.
We provide two simple implementations for you to start with: a naive three-loop implementation similar to shown above and a more cache-efficient blocked implementation.
The necessary files are in matmul.tar. Icluded are the following:
|
The vendor's code uses the vendor implementation of BLAS (on Edison (Cray XC30), you should compare with the Cray LibSci implementation of dgemm). See instructions in the Makefile on compiling the code with different compilers available on Edison. A detailed help on using the programming environment can be found on the NERSC pages.
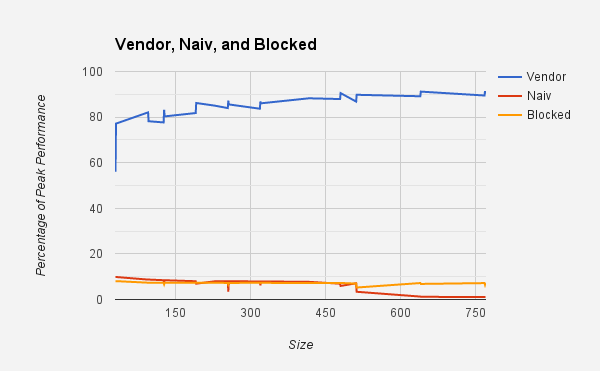
The target processor is a 12-core Intel Ivy Bridge running at 2.4 GHz, yielding 4 double-precision (ie, 64-bit) flops per pipeline * 2 pipelines * 2.4 GHz = 19.2 Gflops/s. The following graph shows the performance of the supplied naive and blocked implementations using the GNU C compiler (gcc). You may use any compiler available. We recommend starting with the GNU C compiler. If you use a compiler other than gcc, you will have to change the provided Makefile, which uses gcc-specific flags. Note that the default compiler, every time you open a new terminal, is Intel - you will have to type module swap PrgEnv-intel PrgEnv-gnu to switch to, eg, GNU compilers. You can type module list to see which compiler wrapper you have loaded. The vendor implementation uses the Cray LibSci, the default vendor-tuned BLAS. The Cray compiler wrappers automatically handle all the linking. If you wish to compare with other BLAS implementations, check the NERSC documentation. Your goal is to get a high performance and understand how you did that. Ideally, you would match or surpass the performance of the BLAS implementation (please check how fast it is), but this is not required.
You may not use compiler flags that automatically detect dgemm kernels and replace them with BLAS calls, i.e. Intel's -matmul flag.
- GNU C provides many extensions, which include intrinsics for vector (SIMD) instructions and data alignment. (Other compilers may have different interfaces.)
- Ideally your compiler injects the appropriate intrinsics into your code automatically (eg, automatic vectorization and/or automatic data alignment). gcc's auto-vectorizer reports diagnostics that may help you identify if manual vectorization is required.
- To manually vectorize, you must add compiler intrinsics to your code. See here for how to use 256 bit vectors.
- Consult your compiler's documentation.
You may assume that A and B do not alias C; however, A and B may alias each other. It is semantically correct to qualify C (the last argument to square_dgemm) with the C99 restrict keyword. There is a lot online about restrict and pointer-aliasing - this is a good article to start with.
The matrices are all stored in column-major order, i.e. Ci,j == C(i,j) == C[(i-1)+(j-1)*n], for i=1:n, where n is the number of rows in C. Note that we use 1-based indexing when using mathematical symbols and MATLAB index notation (parentheses), and 0-based indexing when using C index notation (square brackets).
A version of the source code that runs on IMADA terminal machines can be found in in matmul-imada.tar. Note that the code for the IMADA pool machines does of course not use the Cray library, but the standard ATLAS library (not specifically tuned towards the IMADA HP terminal machines, having an Intel Core i5, 3.20GHz).
Useful References
Technical Publications:
- Goto, K., and van de Geijn, R. A. 2008. Anatomy of High-Performance Matrix Multiplication, ACM Transactions on Mathematical Software 34, 3, Article 12.
- Chellappa, S., Franchetti, F., and Püschel, M. 2008. How To Write Fast Numerical Code: A Small Introduction, Lecture Notes in Computer Science 5235, 196–259.
- Bilmes, et al. The PHiPAC (Portable High Performance ANSI C) Page for BLAS3 Compatible Fast Matrix Matrix Multiply.
- Lam, M. S., Rothberg, E. E, and Wolf, M. E. 1991. The Cache Performance and Optimization of Blocked Algorithms, ASPLOS'91, 63–74.
Technical Documentation:
- Each core on edison nodes has its own L1 and L2 caches, with 64 KB (32 KB instruction cache, 32 KB data) and 256 KB, respectively; A 30-MB L3 cache shared between 12 cores on the "Ivy Bridge" processor. See the NERSC webpages and Specification of the compute nodes of Edison (Intel® Xeon® Processor E5-2695 v2).
- PGI, Pathscale, and Cray Compilers on Edison
- Running Jobs on Edison
- Using BLAS/LAPACK/ATLAS on Linux based systems
- Automatically Tuned Linear Algebra Software (ATLAS)
- A brief API reference for the ANSI/ISO C interface to the BLAS.
You are also welcome to learn from the source code of state-of-art BLAS implementations such as GotoBLAS and ATLAS. However, you should not reuse those codes in your submission.
Report / Submission
The assignment can be done alone, or in groups of two people. The submission of the source code (at least an optimized version of dgemm.c or dgemm.cpp and a Makefile has to be supplemented by a write-up (max. 10 pages), one write-up for the whole group). The write-up should contain
- the names and email addresses of the people in your group (front page),
- the following table on your front page, that indicates the usage of SSE:
\begin{tabular}{ll}
Aligned SSE: & yes/no \\
Unaligned SSE: & yes/no \\
\end{tabular}where aligned SSE means that the data was loaded using unaligned vector
loads (_mm_storeu_pd, mm_loadu_pd), as opposed to aligned loads
(_mm_store_pd, mm_load_pd).
- a statement of who contributed with what,
- the optimizations used or attempted,
- the results of those optimizations,
- the reason for any odd behavior (e.g., dips) in performance, and
- how the same kernel performs on a different hardware platform (and a small discussion of that),
- a figure fhat shows the performace similar than the figure above, but using all matrix sizes 2, 3, 4, ..., 769 for a single processor core of NERSC Edison cluster (y-axis should say percentage of peak performace),
- a figure fhat shows the performace similar than the figure above, but using all matrix sizes 2, 3, 4, ..., 769 for a single processor of another system of your choice (using the same kernel). Explain how you dertermined the peak performace, and again, the y-axis should say percentage of peak performace.
For the last requirement, you may run your optimized implementation on another NERSC cluster, on your laptop, on one of the IMADA pool machines, etc. Note that on the IMADA pool machines ATLAS (Automatically Tuned Linear Algebra Software) is installed as a standard library as well as tuned towards the Intel 3.2GHz machines (see above) .
Passing this assignment will mostly depend on two factors:
- performance sustained by your codes on Edison,
- explanations of the performance features you observed.
For submitting the report, and the sources proceed as follows: Create the following directory structure one of IMADAs pool machines (of course you have to copy the data from the NERSC filesystem to the IMADA system before doing that)
mandatory1/ mandatory1/report/ mandatory1/sources/edison/ mandatory1/sources/other/ mandatory1/results/edison/ mandatory1/results/other/
Put your report, sources, and results in the corresponding
directory. The source directory for the Edison Cluster should
include all files that are necessary to produce an executable code on
the NERSC resources (for example there has to be
a dgemm.cpp and a Makefile in any case). It
should be possible to copy the complete data to the file system of the
Edison cluster and to produce executable files via a
simple make command. The report direcory and
the sources directories should not contain more than 10MB
of data. Each of the two result directories should contain a file
called performance. These two files have to have the
format of the output from the benchmark program (see source code). Use
all matrix sizes 2, 3, 4, ..., 768, 769. The file(s) should look
identical (of course the value behind Mflops/s changes) to the
following:
Size: 2 Mflop/s: 1466.75 Size: 3 Mflop/s: 1515.29 ... Size: 768 Mflop/s: 433.163 Size: 769 Mflop/s: 397.745
Submission deadline and procedure.
Please create a zip file of the report, the sources, and the result directory: Change into the directory containing the directory mandatory1 and type
zip -r mandatory1 mandatory1
This will create a file called mandatory1.zip. This is the file you should submit via blackboard. The zip file should not be larger than 10 MB.
Additionally, you shall hand in a printout of your report and of your source code..
The strict deadline for submission (electronically via blackboard) is October 20, 10:00. Note that the second mandatory assignment will be announced October 4th already. As there is no teaching in week 42, I allow to hand in the printout of the report and the code until October 23, 15:00 (in the lecture), however, the electronical version has to be submitted until October 20th, 10:00. You can put the printout in my mailbox, bring it to my office (slide it under the door if locked), or give it to me in the lecture.
Frequently Asked Questions (FAQ)
- Are we allowed to work in groups?
- What performance should we achieve?
Yes, but you should clearly state who contributed with what in the report. The mandatory assignments are part of your final exam, i.e., you have to fully understand all parts of what you have done.
Besides a good quality report you should aim at 30% of theoretical peak performance.